Here we describe the Cancer Cell Line Encyclopedia (CCLE). The CCLE dataset is available from http://www.broadinstitute.org/ccle publicly.
In order to characterize cancer cell lines in depth and accurately, The Cancer Cell Line Encyclopedia (CCLE) was completed in 2012 by teams from Broad Institute, Dana-Farber Cancer Institute and Novartis Biomedical Research Institute in the United States[1]. They sequenced 947 human cancer cell lines from more than 30 tissue sources, integrating genetic information such as DNA mutations, gene expression and chromosome copy number. In the seven years since it was open to the public, the database has become one of the standard reference databases for cancer genomics: the original paper has been cited approximately 6,000 times to date.
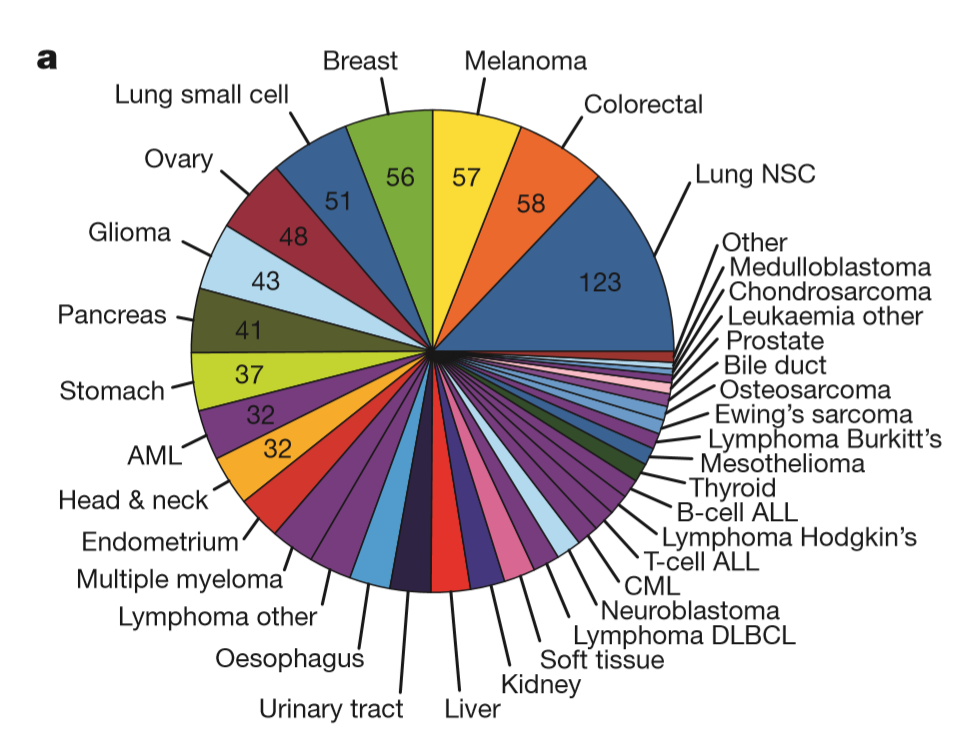
Overview of 2012 CCLE database[1].
With the in-depth development of multi-omics sequencing technology and cancer precision medicine, CCLE database is constantly updated in terms of the number of cancer cells and the dimension of sequencing information. On May 9, 2019, CCLE project team published a long article next-generation characterization of the Cancer Cell Line Encyclopedia on Nature, reporting a major update of the Encyclopedia of Cancer Cell series[2].
In addition to previously available information on DNA mutations, gene expression, and chromosome copy number, CCLE researchers performed a new large-scale analysis of more than 1,000 cancer cell lines (below):
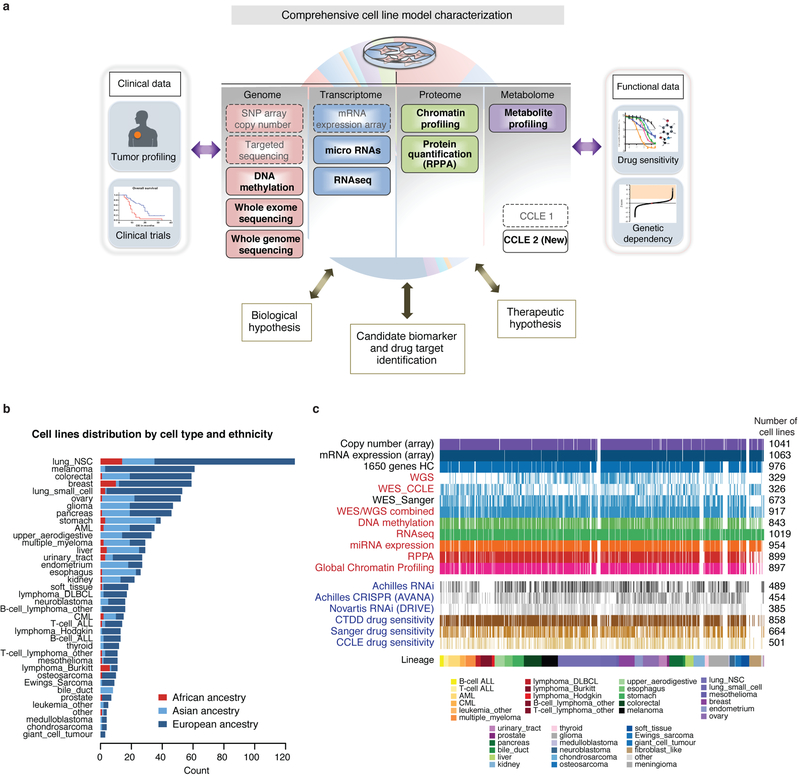
Overview of 2019 CCLE cell lines and datasets.[2].
a, The existing and new CCLE datasets as indicated are depicted. b, Distribution of cell lines by lineage and ancestry across CCLE. c, Visual representation of the number of cell lines in each dataset. New CCLE datasets are shown in red. Functional genomics datasets are shown in blue.
1) Quantitative analysis of 213 proteins in 899 cancer cell lines based on reverse Phase protein Array (RPPA) was performed.
2) Quantitative analysis of gene expression and variable splicing was performed for 1019 cancer cell lines based on RNA-sequencing;
3) 326 cancer cell lines were subjected to whole-exomesequencing (WES) and 329 cancer cell lines were subjected to whole-genome sequencing;
4) Quantitative analysis of promoter methylation levels based on reduced representation Bisulfide sequencing (RRBS) was performed for 843 cancer cell lines.
5) miRNA expression was quantitatively analyzed in 954 cancer cell lines.
6) The histone modification levels of 897 cancer cell lines were quantitatively analyzed.
7) Quantitative analysis of 225 metabolites in 928 cancer cell lines was performed.
As characterization of cell lines at the level of nucleic acids reached new levels of completeness they continued to strive towards an understanding of the protein content of cell lines. The vast majority of therapeutics act by interrupting or altering protein function and with the growing interested in antibody-drug conjugates, antibody mediated cellular cytotoxicity (ADCC), and CAR-T cells all directed at surface proteins they sought to try and define the CCLE proteome through mass spectrometry.
To this end, the Gygi lab performed Tandem-mass tagging mass spectrometry to quantify the abundance of proteins in whole cell extracts derived from 375 of the CCLE cell lines. In addition, serine/threonine phosphorylation events were quantified by cxxxxx. In collaboration with the Carr Mass Spectrometry platform at the Broad Institute tyrosine phosphorylation was quantified in a small set of cell lines under conditions of distinct therapeutic perturbations. The first of these data sets has been published in [3].